Using the mantel test to compare ecological matrices using the vegan R package (CC211)
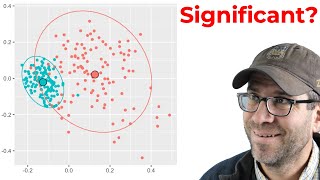
The mantel test is useful for comparing distances matrices and is straightforward to do with the mantel function from the vegan R package. In this Code Club, Pat will use the Mantel test to compare bin-based and phylogenetic beta diversity matrices (weighted and unweighted UniFrac) and to compare membership and structure based metrics of beta diversity. What correlates with what? He'll answer this question using data generated with mothur and R functions from vegan, dplyr, and ggplot2.
You can find my blog post for this episode at https://www.riffomonas.org/code_club/2022-05-09-unifrac-mantel. The data were generated in our Kozich et al. 2013 paper (http://doi.org/10.1128/AEM.01043-13) using samples from the Schloss et al. 2012 paper (http://doi.org/10.4161/gmic.21008).
#microbiome #tidyverse #vegan #R #Rstudio #Rstats #ggplot2
Want more practice on the concepts covered in Code Club? You can sign up for my weekly newsletter at https://shop.riffomonas.org/youtube to get practice problems, tips, and insights.
If you're interested in taking an upcoming 3 day R workshop be sure to check out our schedule at https://riffomonas.org/workshops/
You can also find complete tutorials for learning R with the tidyverse using...
Microbial ecology data: https://www.riffomonas.org/minimalR/
General data: https://www.riffomonas.org/generalR/
0:00 Introduction
4:09 Generating Bray-Curtis and Jaccard distances
7:00 Importing Unweighted and Weighted Unifrac distances
11:38 Graphically comparing distance methods
16:14 Using mantel test to compare distance methods